Repeated evolution in natural populations
When does genome evolution repeat itself?
Evolution is driven by a deterministic forces like directional selection but also by chance, what brings an intriguing question about its predictability. Because evolution is a historical process, we have limited possibilities how to experimentally test for such question in its full complexity, i.e. in natural environments.
Distinct lineages, both close populations and distant species, that independently evolved under similar conditions (i.e. underwent repeated adaptation) represent useful study systems. Knowing how predictable is parallel evolution in such natural experiments should inform about how deterministic is evolution in general and provide insights into predictive evolution of crops, pathogens or species under climate change.
We have found that gene reuse is shaped by the extent of variation shared among populations within a genus Arabidopsis (see the page bottom). Yet, the mechanisms acting at greater evolutionary distances remain controversial. Now, we are extending the divergence scale to the entire Brassicaeae family spanning 25 M years of divergence. Leveraging independent colonizations of toxic serpentine soil (ConAdapt project) and replicated transitions to polyploidy (PolyAdapt project) in many Brassicaceae species, we aim at systematic assessment of the factors underlying repeated genome evolution and thus evolutionary predictability.

These are collaborative projects involving many colleagues, in particular Levi Yant (SLU Uppsala), Sam Yeaman (U Calgary), Korbinian Schneeberger (LMU Munich), Tomica Mišljenović (U Belgrade), Panayiotis Dimitrakopoulos (Aegean U, Lesbos), Terezie Mandáková and Martin Lysak (Ceitec Brno), Stanislav Španiel (SAS Bratislava) and Matyáš Fendrych (CAS, Prague)
ConAdapt project (2023-2027)
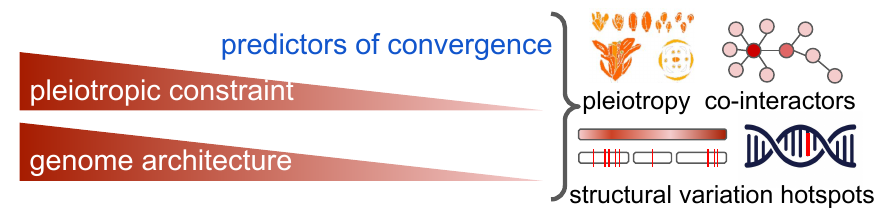
We investigate the probability of repeated adaptive evolution on genome level, by studying genomic variation of Brassicaceae species that independently adapted to the same selective pressure. The project is funded by a five-years high-risk Junior Research Talent (JuniorStar) funding scheme of the Czech Science Foundation.
We combine population and structural genomics with transcriptomic and reverse genetic validations to characterize the factors determining genomic hotspots of convergence and test the hypothesis of their varying importance with divergence. Our model system of adaptation are naturally toxic serpentine soils, detailed at the link below.


In the first phase of the project, our main focus is on sampling, experimental cultivation and genome sequencing of natural diversity of the target groups in order to identify candidate loci for serpentine adaptation. We are also progressing with new genome assemblies of the representatives of the following genera: Aethionema, Alyssum, Aubrieta, Bornmuellera, Cardamine, Draba, Erysimum, Isatis, Noccaea, Odontarrhena and Rorippa. Please, contact us if you are interested in collaboration or further use of these resources.

PolyAdapt project (2026-2028)
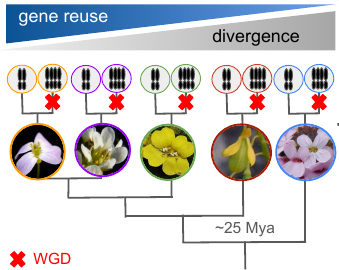
Whole genome duplication (WGD) is a dramatic mutation that provides evolutionary opportunity but also imposes severe stresses associated with cell division and homeostasis, to which nascent polyploids have to adapt. In this project funded by the Czech Science Foundation, we test if genome architecture or functional constraints play the key role in repeated natural adaptation to WGD. Based on a systematic sampling of 13 Brassicaceae species varying in ploidy, we identify repeated footprints of post-WGD selection by comparing genomes of established polyploids with their sister diploid populations. Then we will leverage extensive genetic resources available for this model family to infer key genomic predictors for adaptive gene reuse.
Repeated evolution in alpine Arabidopsis (2017-2020)
We studied the probability of parallel adaptive evolution on genome level in seven independent alpine lineages in Arabidopsis.
First, by combining transplant experiments, genome resequencing and transcritptomics, we revealed set of candidate genes and functional pathways that have been repeatedly recruited for alpine adaptation.
Using this unique seven-fold replicated system, we demonstrated that the probability of gene reuse decreased with divergence between compared lineages, likely because less alleles are shared among distantly related lineages. This means that more divergent lineages have less variation available for gene reuse and, subsequently, are less likely to evolve parallel genetic solutions to similar challenges.
This research if further followed up by the team of Magdalena Bohutínská by testing phenotypic effects and adaptive significance of the repeated candidate loci.
In general, our results suggest that genetic basis of adaptive evolution could be better predictable in closely related lineages with shared pool of adaptive alleles while it becomes subject of change in distantly related lineages.



